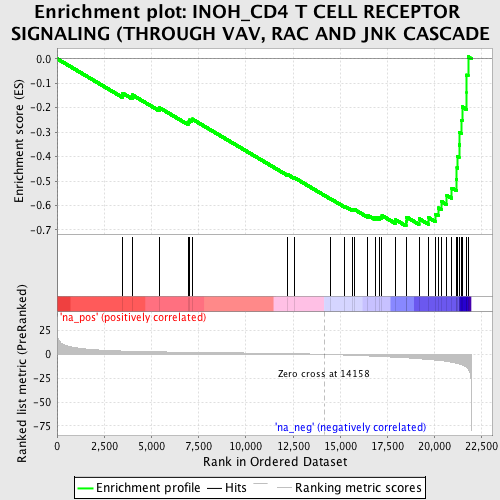
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_CD4 T CELL RECEPTOR SIGNALING (THROUGH VAV, RAC AND JNK CASCADE |
| Enrichment Score (ES) | -0.6838595 |
| Normalized Enrichment Score (NES) | -2.229776 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOSL1 | 3479 | 3.447 | -0.1411 | No | ||
| 2 | BTK | 3984 | 3.157 | -0.1479 | No | ||
| 3 | MAPK10 | 5422 | 2.586 | -0.2002 | No | ||
| 4 | FOSB | 6942 | 2.132 | -0.2586 | No | ||
| 5 | RAC1 | 7002 | 2.119 | -0.2504 | No | ||
| 6 | RAC3 | 7147 | 2.079 | -0.2463 | No | ||
| 7 | FOSL2 | 12202 | 0.782 | -0.4729 | No | ||
| 8 | JUN | 12565 | 0.666 | -0.4860 | No | ||
| 9 | ITPR3 | 14478 | -0.207 | -0.5722 | No | ||
| 10 | SRC | 15241 | -0.705 | -0.6034 | No | ||
| 11 | LAT | 15646 | -0.971 | -0.6168 | No | ||
| 12 | MAP2K1 | 15741 | -1.040 | -0.6158 | No | ||
| 13 | ZAP70 | 16444 | -1.608 | -0.6396 | No | ||
| 14 | RAC2 | 16856 | -1.992 | -0.6481 | No | ||
| 15 | TYK2 | 17095 | -2.213 | -0.6476 | No | ||
| 16 | MAPK9 | 17214 | -2.310 | -0.6411 | No | ||
| 17 | JAK3 | 17929 | -3.030 | -0.6582 | No | ||
| 18 | SYK | 18493 | -3.653 | -0.6651 | Yes | ||
| 19 | JAK2 | 18520 | -3.677 | -0.6474 | Yes | ||
| 20 | FOS | 19178 | -4.546 | -0.6541 | Yes | ||
| 21 | JAK1 | 19675 | -5.293 | -0.6495 | Yes | ||
| 22 | VAV1 | 20066 | -6.018 | -0.6365 | Yes | ||
| 23 | PLCG1 | 20195 | -6.259 | -0.6102 | Yes | ||
| 24 | MAP2K2 | 20359 | -6.618 | -0.5836 | Yes | ||
| 25 | PTK2 | 20652 | -7.365 | -0.5592 | Yes | ||
| 26 | PRKCQ | 20917 | -8.226 | -0.5290 | Yes | ||
| 27 | ITPR1 | 21161 | -9.189 | -0.4929 | Yes | ||
| 28 | CD4 | 21188 | -9.300 | -0.4464 | Yes | ||
| 29 | MAPK8 | 21208 | -9.408 | -0.3990 | Yes | ||
| 30 | FYN | 21301 | -9.911 | -0.3523 | Yes | ||
| 31 | ITK | 21334 | -10.083 | -0.3020 | Yes | ||
| 32 | CSK | 21416 | -10.654 | -0.2510 | Yes | ||
| 33 | MAP2K4 | 21466 | -10.991 | -0.1968 | Yes | ||
| 34 | PTK2B | 21693 | -13.678 | -0.1370 | Yes | ||
| 35 | TEC | 21697 | -13.757 | -0.0665 | Yes | ||
| 36 | LCK | 21778 | -15.166 | 0.0077 | Yes |